MetScape Plugin for Cytoscape
Please note that this version of MetScape is out of date. For the current version, go to MetScape 3
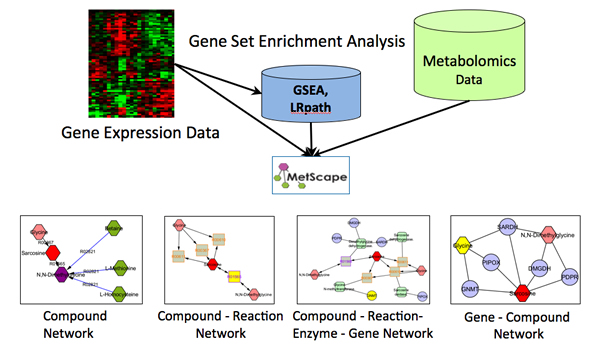
The MetScape Plugin for Cytoscape provides a bioinformatics framework for the visualization and interpretation of metabolomic and expression profiling data in the context of human metabolism. It allows users to build and analyze networks of genes and compounds, identify enriched pathways from expression profiling data, and visualize changes in metabolite data. Gene expression and/or compound concentration data can be loaded from file(s) (in CSV, TSV, or Excel formats), or the user can directly enter individual compounds/genes (using KEGG compound IDs, or Entrez Gene IDs) to build metabolic networks without loading a file. MetScape uses an internal relational database stored at NCIBI that integrates data from KEGG and EHMN.
Download the latest version of MetScape | Launch Cytoscape w/ MetScape 2 via Java Web Start
Download sample input files: concepts.xlsx, genes.xlsx, metabolites.xlsx
Download beta version of MetDisease
Project Leaders
Developers
Tim Hull
Glenn Tarcea
Terry Weymouth
Acknowledgments
Charles Burant
Barbara Mirel
Maureen Sartor
Alex Ade
Yongsheng Bai
Jim Cavalcoli
Jing Gao
Rich McEachin
Jean Song
Marci Brandenburg
Alex Terzian
Paul Trombley
H.V. Jagadish
Gilbert S. Omenn
Brian Athey
Please report any problems to metscape-help@umich.edu
Funding
NCIBI is funded by the National Institutes of Health: Grant #U54 DA021519. PI: Brian Athey
Pilot and feasibility grant from Michigan Diabetes Research and Training Center. PI : Alla Karnovsky
Cite Metscape
