Help & Support
Download MetScape 2.0 User Manual | Watch MetScape Online Video Tutorial
Download MetScape 3.0.2 User Manual
For additional help, please contact metscape-help@umich.edu
Input File Formats
MetScape 2 allows users to upload three types of files – compound file, gene file and concept file. Each type is optional, e.g. you can upload only compounds, only genes, only concepts, or any combination of the above.
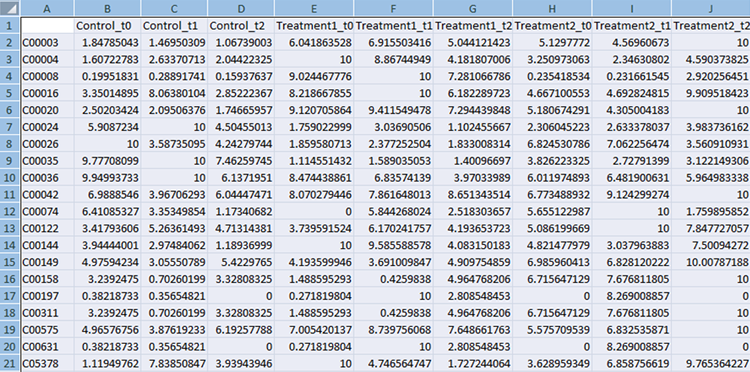
Compound File
The first column should always contain KEGG compound IDs. The subsequent columns should contain normalized experimental measurements. If the experiment has several time points they should be listed in the adjacent columns, as shown in the example below, e.g. control t0, control t1, control t2 etc. The top row contains column headings that will be used to label the data.
Gene File
The first row should contain column headings that label the data. The first column should always contain Entrez Gene IDs. The second column should contain p-values for differentially expressed genes (e.g. from test). The third column can contain either log fold change or fold change values.
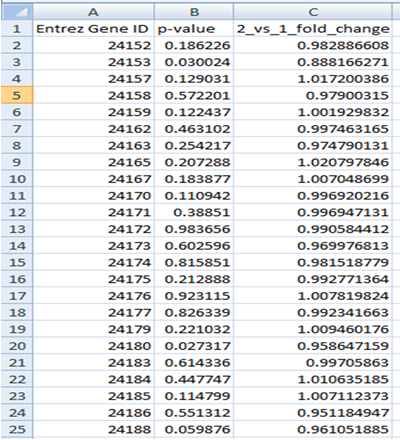
Gene file can contain all genes (e.g. from microarray experiment ) or significant genes only.
Please note that the list of all genes is required in order to run LRpath (see complete user manual for details).
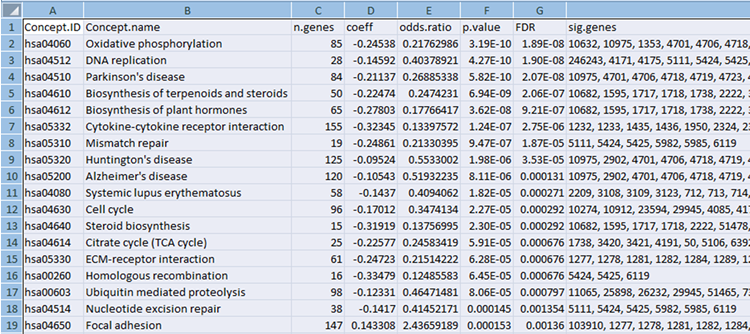
Concept File
Concept file can be generated by gene set enrichment analysis tool such as LRpath or GSEA from gene expression data.
Gene set enrichment testing is an approach used to test for predefined biologically-relevant gene sets that contain more significant genes from an experimental dataset than expected by chance.
- GSEA (Subramanian at al., Proc. Natl. Acad. Sci. USA, 2005, 102, 15545-15550)
- LRpath (Sartor et al., 2009, Bioinformatics. 2009, 25(2):211-7.
Instead of uploading concept file you can run LRpath directly from MetScape. See complete user manual for details.